
These nonprotein-coding RNAs (ncRNAs) come in many different forms, and they have been shown to have a variety of functions within the cell, influencing processes such as gene expression, mRNA splicing, and transport, just as a few examples. With the many advances in genome-wide sequencing, it has been discovered that much more of the genome is transcribed into RNA than previously appreciated.

Methodology and first applications to the phage T4 DNA replication system.
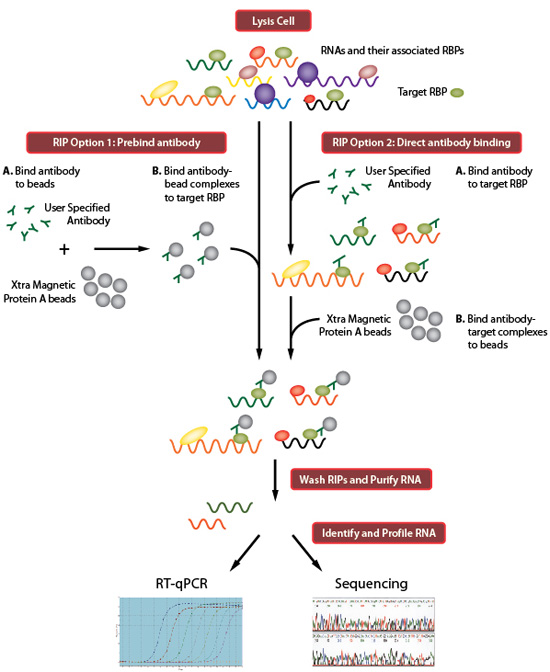
ICLIP EXPERIMENTS PROTOCOLS RNA IP CODE
Yeo GW, Coufal NG, Liang TY et al (2009) An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Sugimoto Y, Konig J, Hussain S et al (2012) Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions, Genome Biol 13:R67 Langmead B, Trapnell C, Pop M et al (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol 10:R25 Yao C, Biesinger J, Wan J et al (2012) Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Hafner M, Renwick N, Brown M et al (2011) RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. Konig J, Zarnack K, Rot G et al (2010) iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Ule J, Jensen KB, Ruggiu M et al (2003) CLIP identifies Nova-regulated RNA networks in the brain. Gilbert C, Kristjuhan A, Winkler GS et al (2004) Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation.

Tenenbaum SA, Carson CC, Lager PJ et al (2000) Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. RNA 5:1071–1082īrooks SA, Rigby WF (2000) Characterization of the mRNA ligands bound by the RNA binding protein hnRNP A2 utilizing a novel in vivo technique.
Trifillis P, Day N, Kiledjian M (1999) Finding the right RNA: identification of cellular mRNA substrates for RNA-binding proteins. Hieronymus H, Silver PA (2004) A systems view of mRNP biology. Castello A, Fischer B, Eichelbaum K et al (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins.


 0 kommentar(er)
0 kommentar(er)
